
The CIC, home of the Mouse Connectome Project (MCP) seeks to develop a multimodal multiscale connectome and cell-type map of the mammalian brain using advanced tracing, imaging, and computational methods.
Our crossdisciplinary group develops neuroanatomic and neuroinformatic approaches to understand connectivity patterns in both health and disease.
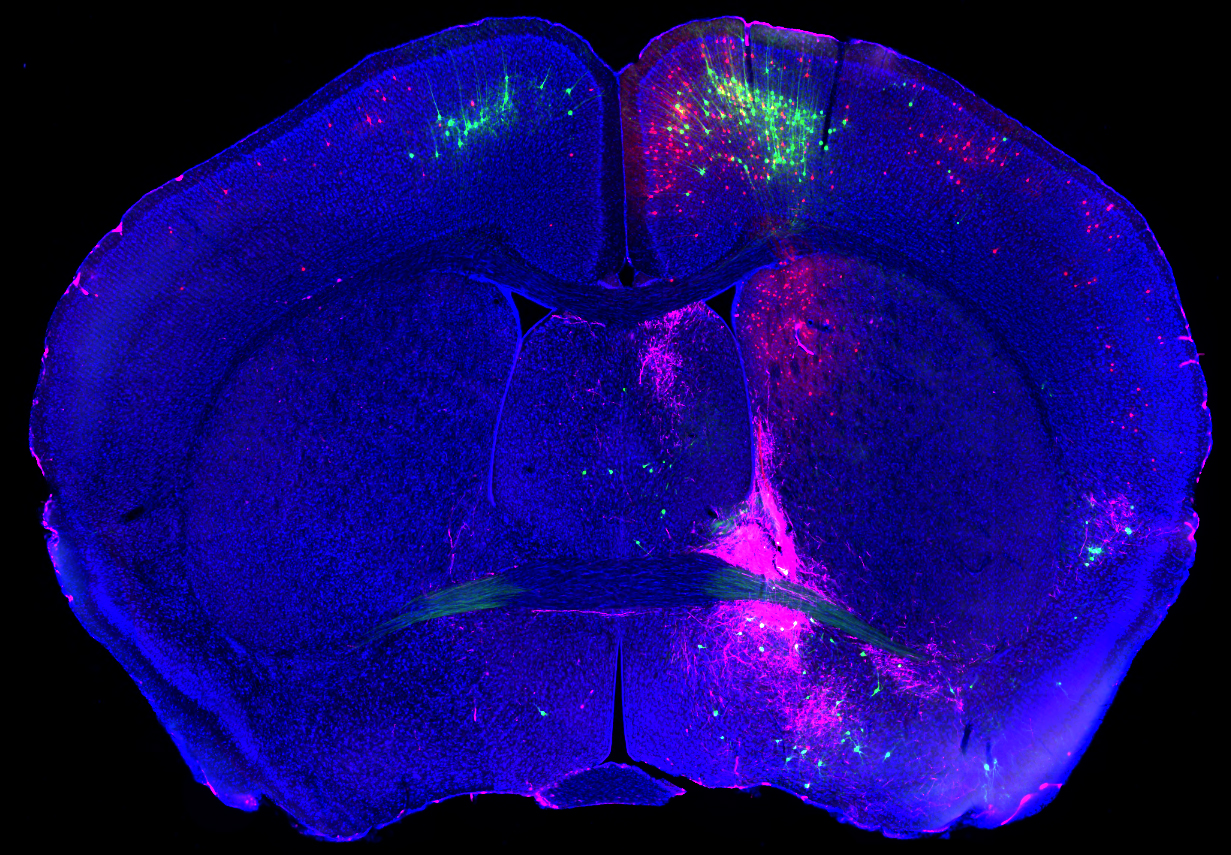
The Mouse Connectome Project (MCP) is an NIH-funded project that aims to create a complete mesoscale connectivity atlas of the C57Black/6 mouse brain and to subsequently generate its global neural networks. This will help researchers gain a better understanding of how different brain structures organize into networks and how they communicate with one another to influence behavior. Multiple fluorescent neural dyes are used to trace all of the connections of the roughly 800 identified structures of the mouse brain. Novel computational neuroinformatics tools, developed through the collaborative efforts of MCP neuroanatomists and computer scientists, are used to quantify and analyze the large-scale datasets, to extract the valuable information embedded in the terabytes of acquired image data.
View Data
The Center for Epigenomics of the Mouse Brain Atlas (CEMBA), will require collecting data from over 100 brain regions covering the entire mouse brain and associating those cell types with anatomical features that may be common to nervous systems generally and therefore subject to similar methods of monitoring and manipulation in other species.
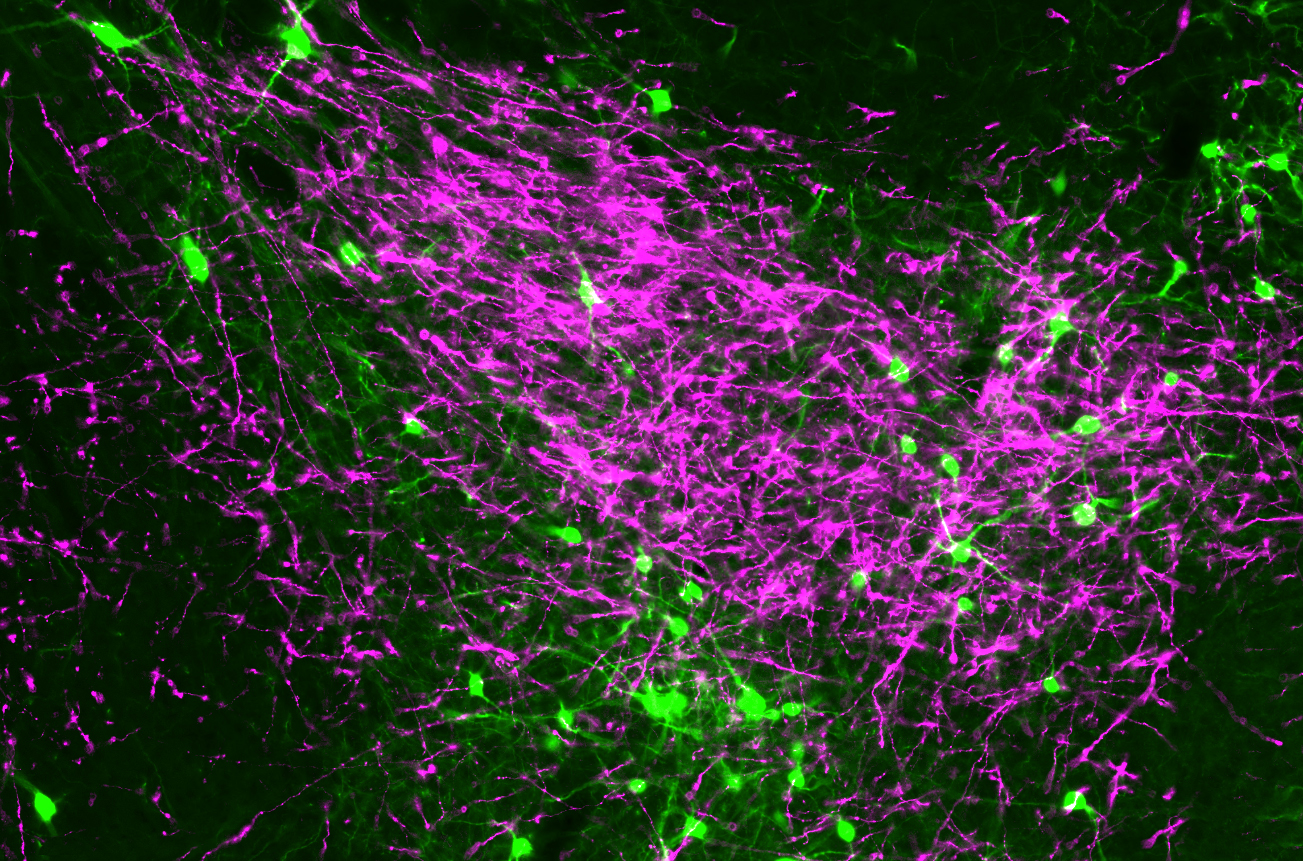
Our U01 project proposes an anatomical characterization of cell types of the mouse brain. A combination of techniques, including viral circuit tracing, CLARITY, and Expansion Microscopy, will be used to reveal cell type anatomy, morphology, axonal trajectories, and monsynaptic input/output organization.

View Data
The mouse brain is composed of about one hundred million highly interconnected neurons. A major component of deciphering the immense complexity of the mammalian brain is to obtain highly detailed and reproducible information at the level if the individual neuron. In this BRAIN Initiative-funded study, the Yang lab will apply a novel mouse genetic tool to sparse-label specific genetically-defined neuronal populations, image at high-resolution, and analyze this large-scale neuronal morphological data (i.e. dendrites) obtained from the mouse brain. The clustering of neurons based on their morphological features could help define novel neuronal cell types and contribute to the BRAIN Initiative's effort to generate a reference mouse brain cell atlas.
View Data
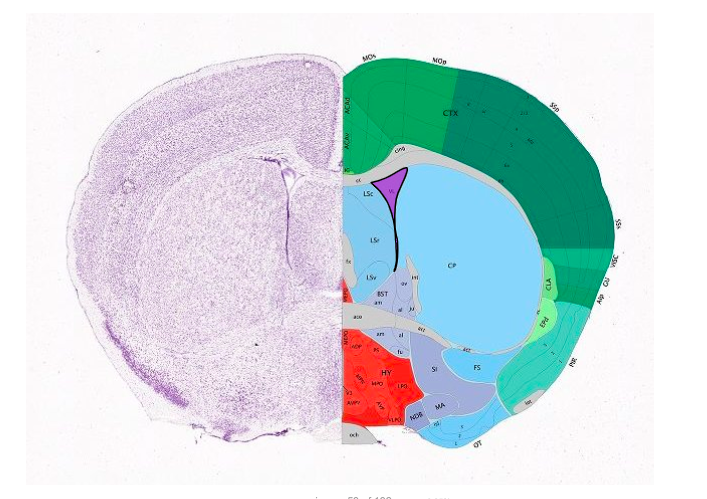
The Allen Reference Atlas is a digital, high-resolution anatomic reference atlas of the C57Black/6J male mouse, created by Hong-Wei Dong at the Allen Instititue for Brain Science.

Our in-house software created to reliably and efficiently register and annotate large-scale 2D connectivity hisology data.
View Data
The dashboard page of the data repository of all the published cases from CIC group.
View Data
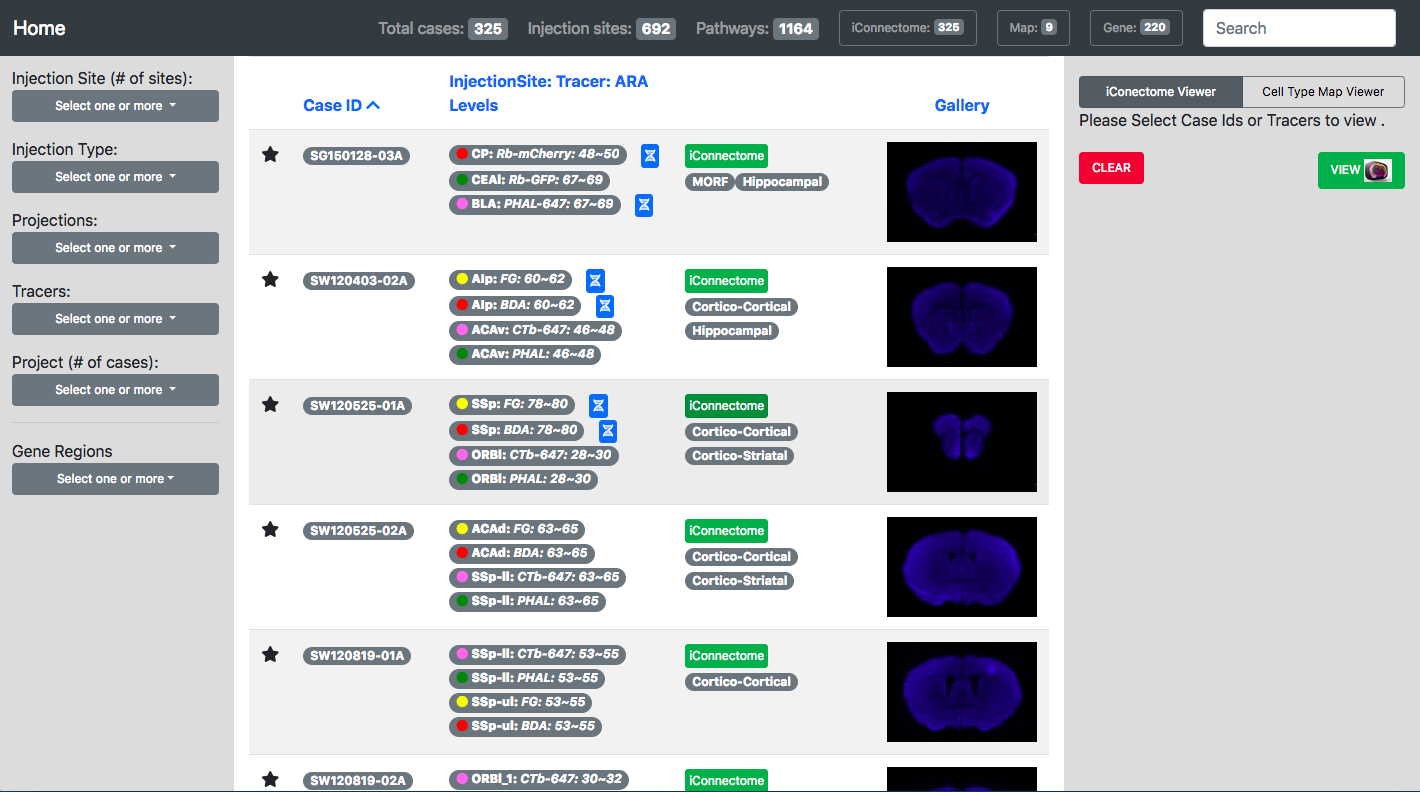
The CIC web portal offers a catalog of neural tracer injection cases, which are updated continuously and eventually will cover the entire brain. Serial sections of each case are available to view at 10x magnification in the interactive iConnectome viewer.
View Data
The CIC web portal also features connectivity maps, which contain graphic reconstructions of the labeling produced by the tracer injections atop a standard mouse brain atlas. These maps provide a fast and easy way to view and compare projection patterns across injections.
View Data
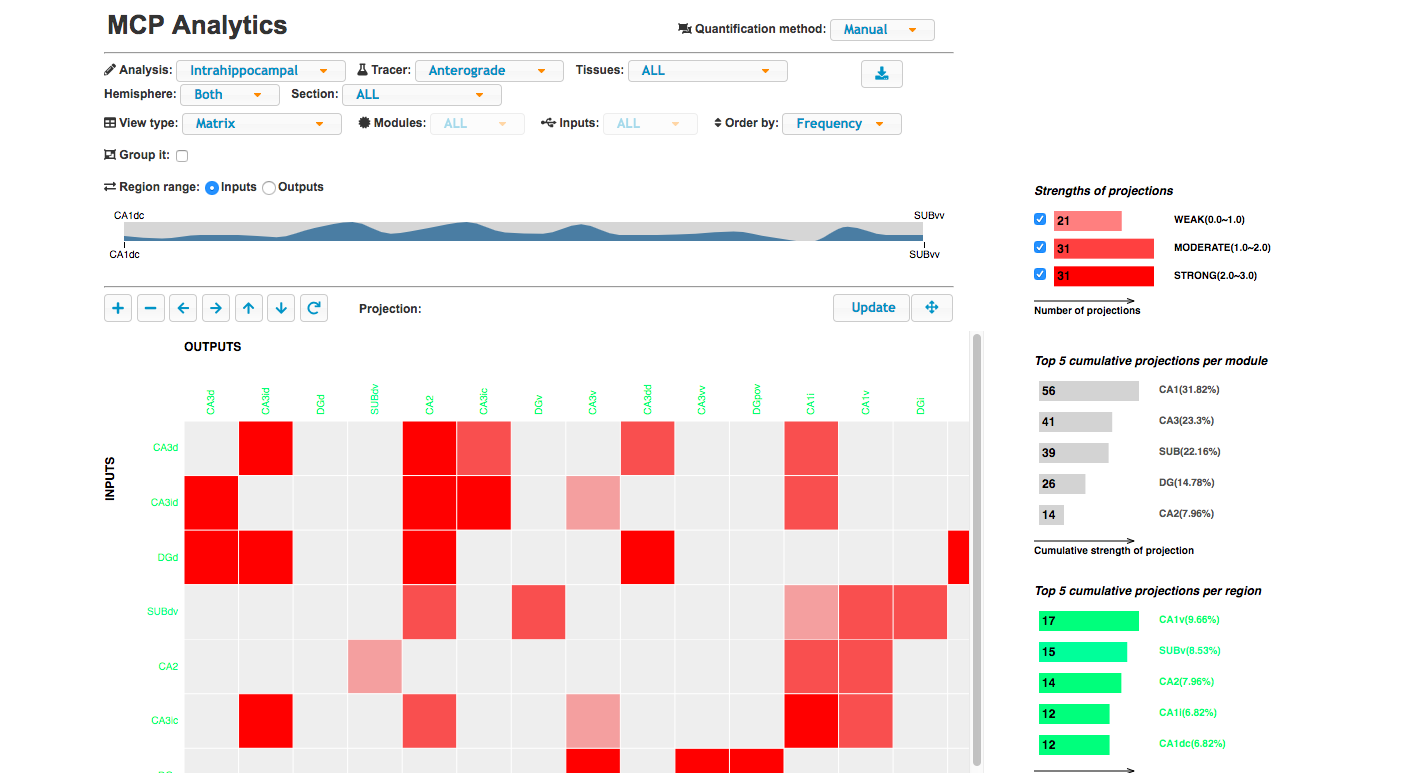
Visualization tool of the projection matrix, ARA tree, bars, and connectivity graph of the hippocampus and cortico-cortico projections.
View Data
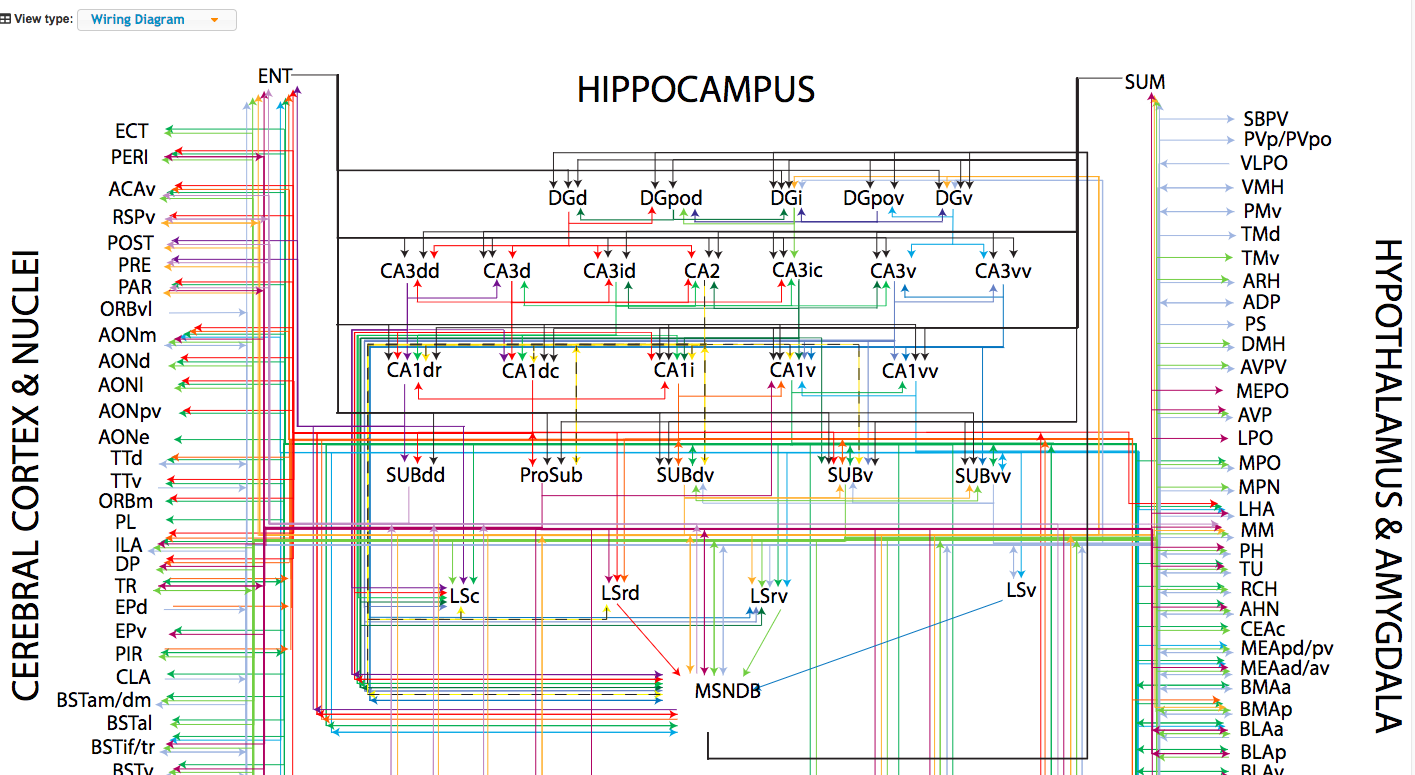
Interactive connectivity wiring digram of the Hippocampus. The most complete hippocampal network wiring diagram to date, which can serve as a foundation for the functional dissection of the hippocampus.

View Data
3D viewer of the Hippocampus Gene Expression Atlas.

View Data
Collection of video archives of 3D mouse connectome data and brain architecture of the cell types.
View Data
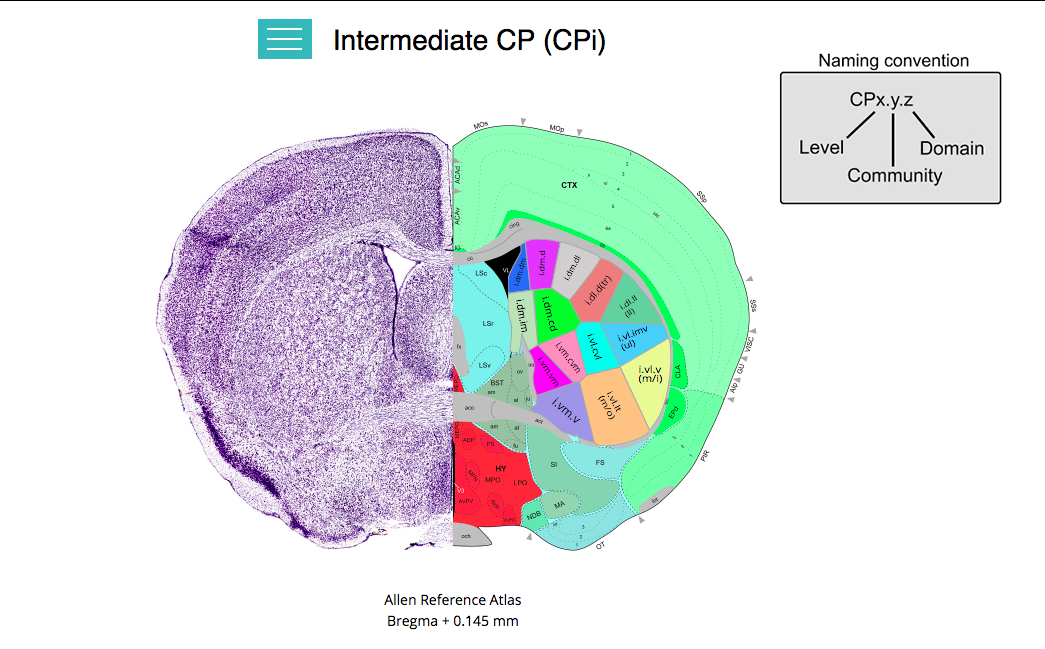
This dynamic map provides a list of all cortical areas that primarily project to each of the domains within the rostral, intermediate, and caudal CP (or dorsal striatum).
View Data
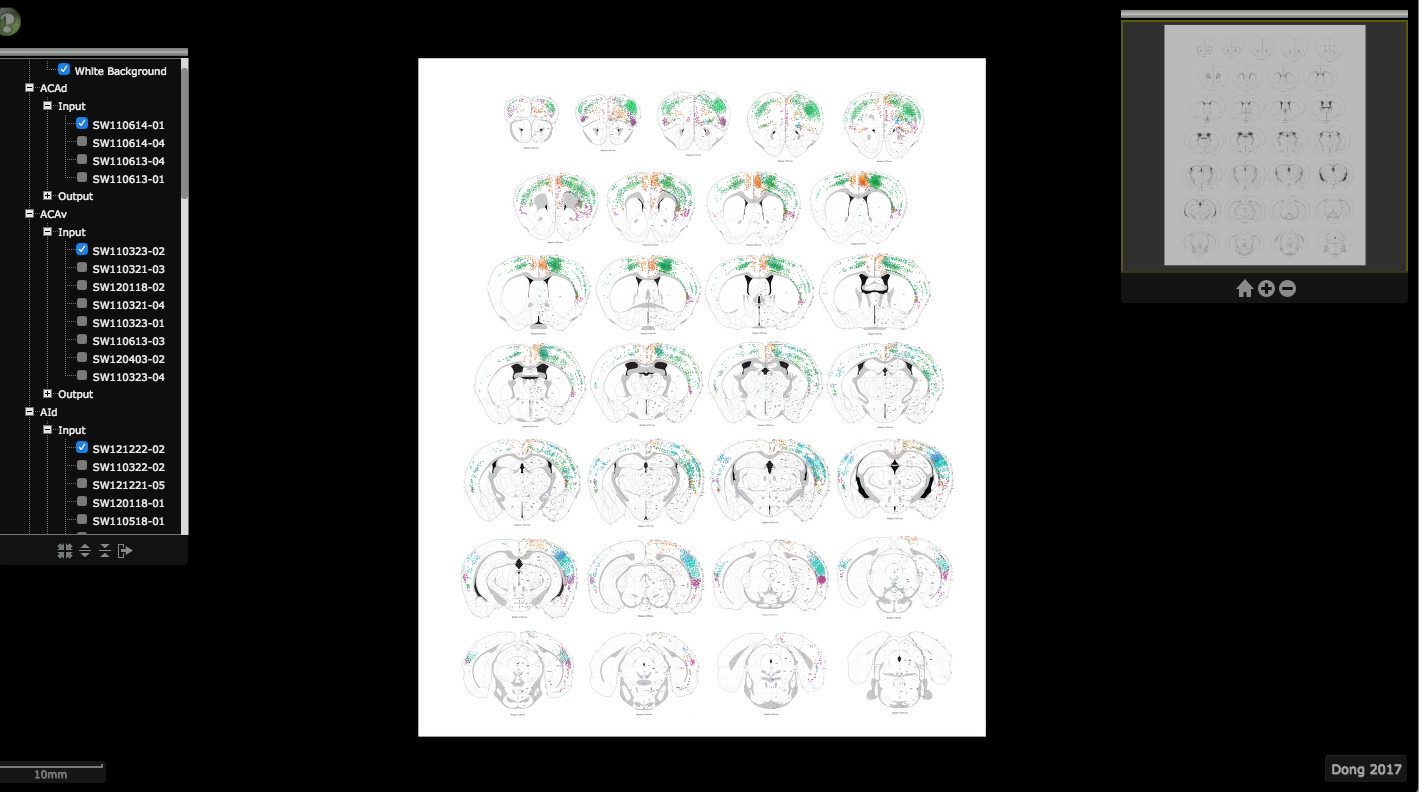
This cortico-cortical map provides an overview of projection patterns for each cortical injection case available in iConnectome. Data were manually reconstructed from raw images--axons appear as shaded regions and retrogradely labeled cells appear as colored dots. A large dot indicates injection site.

View Data
This cortico-striatal map provides reconstructions of projections generated with threshold-based reconstruction algorithm that segments axons labeled by anterograde tracers from tissue background. Only terminal fields within the caudoputamen are depicted. Raw tracer data for all mapped cases are available on iConnectome.
The Mouse Connectome Project (MCP) is an NIH-funded project that aims to create a complete mesoscale connectivity atlas of the C57Black/6 mouse brain and to subsequently generate its global neural networks. This will help researchers gain a better understanding of how different brain structures organize into networks and how they communicate with one another to influence behavior. Multiple fluorescent neural dyes are used to trace all of the connections of the roughly 800 identified structures of the mouse brain. Novel computational neuroinformatics tools, developed through the collaborative efforts of MCP neuroanatomists and computer scientists, are used to quantify and analyze the large-scale datasets, to extract the valuable information embedded in the terabytes of acquired image data.
The MCP team is comprised of a blend of neuroanatomists, computer scientists, and web programmers each with a complementary set of skills that facilitates our multi-disciplinary approach to computational neuroanatomy.
Identifying classes of neural cell types will pave the way for researchers to selectively manipulate circuits and develop cell-type specific therapeutics for neurological diseases. Accurately classifying cell types requires an understanding of multiple neuronal properties including neuromorphology, connectivity, and gene expression. The CIC's multimodal approach will enable creation of the first-ever cell-type map of the mouse brain. Defining cell types is supported by NIH/NIMH RF1MH114112 (Hong-Wei Dong, Long Cai, Li Zhang); NIH/NIMH U01MH114829 (Hong-Wei Dong, Giorgio Ascoli, Byungkook Lim); NIH/NIMH U19MH114821 (Josh Huang, Paola Arlotta); NIH/NIMH U19MH114831 (Joseph Ecker, Edward Callaway). Learn more
Research now shows that disconnections in the brain underlie manifestations of symptoms for disorders including Huntington’s disease, Parkinson’s disease, Alzheimer’s disease, autism spectrum disorders and many others. Therefore, it is important to identify the precise locations of these disconnections to constrain research regarding these disorders to specific regions of the brain. The MCP team applies the connectomics approach to mouse models of different disorders to localize these connectional disruptions. Our recent findings for such pathological connections in Huntington’s disease and autism spectrum disorders is detailed in a recent publication. Go to Connectome data or Article.
The CIC at USC is a collaborative environment hosting scientists from backgrounds ranging from neuroanatomy, to microscopy and computer science, facilitating the production, collection, and analysis of brain-wide connectivity data. The CIC has also established collaborations with institutions outside of USC, as part of an integrated effort to identify and define the neural networks and cell types of the mammalian brain. Academic partners include renowned scientists across the US from a variety of backgrounds: geneticists who create specialized viral circuit tracers, engineers who develop cutting edge imaging technologies, and experts in the field of human disorders including Alzheimer’s and Huntington’s Disease are part of this effort.
